Development of controls for Pocillopora acuta triploidy detection via qPCR
The goal of this notebook post is to explain how I developed the standards we will use for triploidy detection for the Pacu Ploidy project. We are developing a qPCR test to determine the ploidy of P. acuta individuals by detecting copy number of the ITS amplicon after several rounds of qPCR. All DNA used for the method will be normalized by mass, so it should take more cycles to amplify the DNA from the triploid samples because the they have a larger genome size; fewer copies of the amplicon will be present in the same quantity of sample. This process is based off of the geneBlock design protocol (link) used by the Frank Laboratory at Kewalo Marine Lab
Files & Programs
- FASTA file of A18-ITS4 sequences (You & Huang, 2010; link)
- Haploid genome assembly of Pocillopora acuta (link)
- BLAST (command line) version 2.9.0-2
- Amplify4 (link)]
I chose the sequences from You & Huang because they were the only amplicon sequences I could find for any Pocillopora species using the A18 and ITS4 primers we plan to use.
Step 1: BLAST the FASTA file of A18-ITS4 sequences from You and Huang against the P. acuta reference genome
First, index the reference genome. This command is fairly self-expalantory. It makes an index of all the sequences to make the fasta file blast-searchable. The -parse_seqids allows you to keep your original sequence names so yo can reference them later.
makeblastdb -in reference_genome/Pocillopora_acuta_HIv2/Pocillopora_acuta_HIv2.assembly.fasta -input_type fasta -dbtype nucl -parse_seqids -out reference_genome/Pocillopora_acuta_HIv2/
Run BLASTN using the Huang and You FASTA file as the query and the reference genomeas the database. This will find scaffolds that match the ITS2 region. We saved it as an ASN file so that we can convert the results to any format we like with the blast_formatter tool.
blastn -query query/You_and_Huang_2010_Pocillopora_A18S-ITS4_sequences.fasta -db reference_genome/Pocillopora_acuta_HIv2/Pocillopora_acuta_HIv2.assembly.fasta -outfmt 11 -evalue 1e-05 -out blast_results/You_and_Huang_2010_Pocillopora_A18S-ITS4_hits.asn
Format as a TSV file to import into Excel and explore our results
blast_formatter -archive blast_results/You_and_Huang_2010_Pocillopora_A18S-ITS4_hits.asn -outfmt 6 -out blast_results/You_and_Huang_2010_Pocillopora_A18S-ITS4_hits.tsv
Open in Excel and save as an Excel file so that we can filter, sort, highlight, and save our edits.
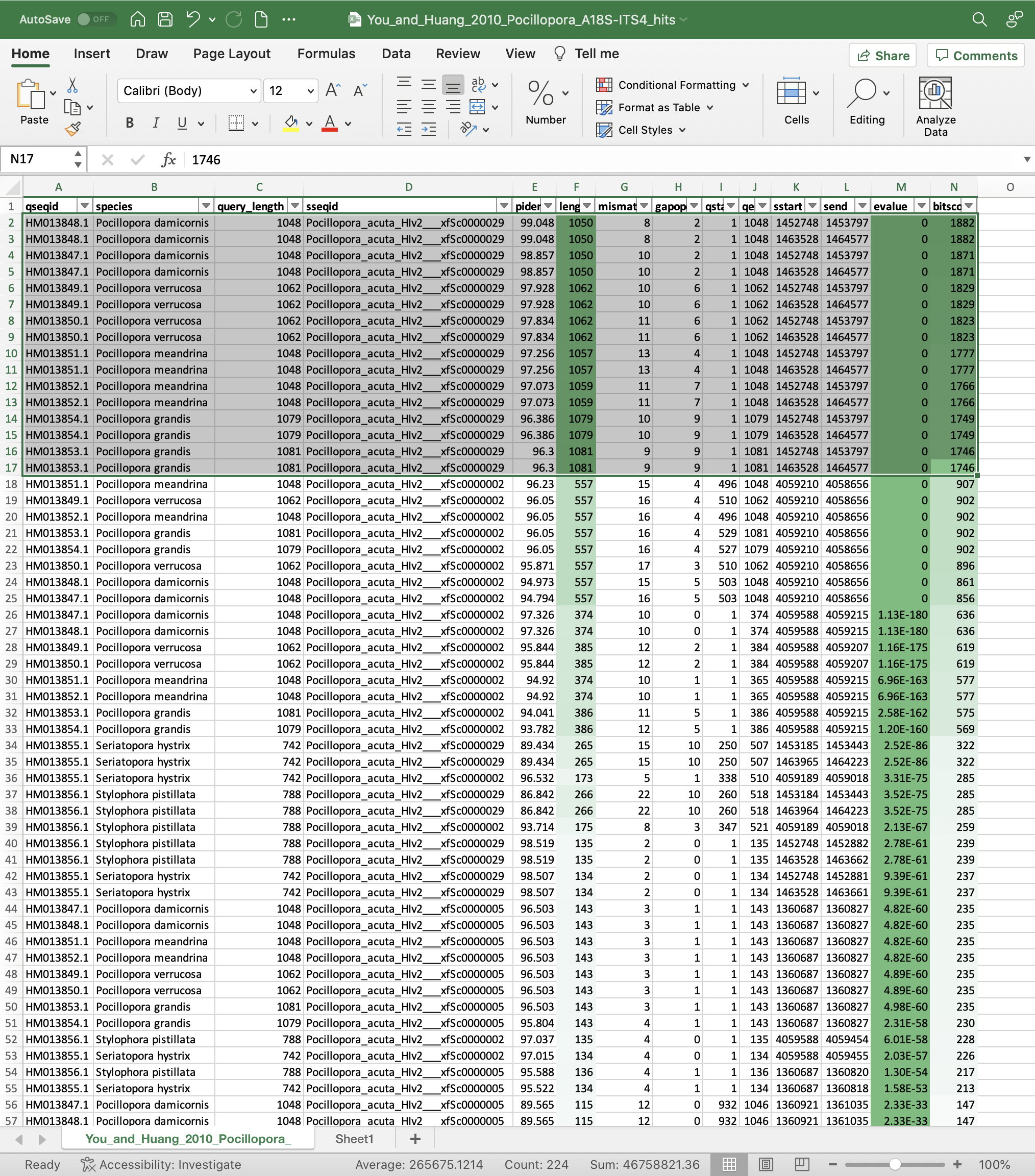
BLAST search results
Our BLAST search results show that the ITS2 region is in scaffold Pocillopora_acuta_HIv2___xfSc0000029 between 1452000 and 1464600 base pairs in our genome assembly. The queried stretch of ~1050-1080 base pairs appears highly conserved among Pocillopora spp., so our primers should be in this general vicinity.
Step 2: Extract the region of interest from the genome assembly
Use blasdcmbdb to extract the region of interest.: Scaffold Pocillopora_acuta_HIv2___xfSc0000029 from 1452000-1464600.
blastdbcmd -entry Pocillopora_acuta_HIv2___xfSc0000029 -db reference_genome/Pocillopora_acuta_HIv2/Pocillopora_acuta_HIv2.assembly.fasta -target_only -range 1452000-1464600 > blast_results/Pocillopora_acuta_HIv2___xfSc0000029_1452000-1464600.A18S-ITS4.fasta
Extracted sequence:
>Pocillopora_acuta_HIv2___xfSc0000029:1452000-1464600
AGACGAACTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAAAGTTAGAGGATCGAAGACGATCAGATACCGTCCTAGTTCTAACCATAAACGATGCCGACTAGGGATCAGAGGGTGTTATTGGATGACCCCTTTGGCACCTCCACGGGAAACCAAAGTGTTTGGGTTCCGGGGGAAGTATGGTTGCAAAGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACCAGGAGTGGAGCCTGCGGCTTAATTTGACTCAACACGGGGAAACTCACCAGGTCCGGACATAGTGAGGATTGACAGATTGAGAGCTCTTTCTTGATTCTATGGGTGGTGGTGCATGGCCGTTCTTAGTTGGTGGAGTGATTTGTCTGGTTAATTCCGTTAACGAACGAGACCTTGACCTGCTAAATAGTGACGCCATCCTCGGATGGGCGGCGAACTTCTTAGAGGGACTGTTGGTATCCAACCAAAGTCAGGAAGGCAATAACAGGTCTGTGATGCCCTTAGATGTTCTGGGCCGCACGCGCGCTACACTGACGACGTCAGAGAGTGCATCCTTCGCCGAAAGGCGCGGGTAATCTGATAAACGTCGTCGTGCTGGGGATAGATCCTTGCAATTATTGATCTTGAACGAGGAATTCCTAGTAAGCGCGAGTCATCAGCTCGCGTTGATTACGTCCCTGCCCTTTGTACACACCGCCCGTCGCTACTACCGATTGAATGGTTTAGTGAGGCCTCCTGACTGGCGCCGACACTCTGTCTCGTGCAGAGAGTGGGAGGCCGGGAAGTTGTTCAAACTTGATCATTTAGAGGAAGTAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTACCGAAAGAGAGATACGTAGTTTTAGTACTACGAGTTCTTCTACCCTGGGAACCAGTGGAAAAAATTATCATTAGGGGTCGGCTGGTTCCGTTAAAAGGAGAGGGCCGCGAGGCGTCGCTCGGACTGCCGTGCTAGGTGACTTTGGTGGAGATGGATGGAGACTGCTCAGTGGGATCGTAGCGTGACCGTGCGGTGTGGCGTAGGACGCTTCGTGCGCCGCGTCGGTGCCTGCCTACGTTTCTCGTTGTGTGTGTCCTTTTCTCTCTCTCCGTGTCTATCTAGTGCGTGCACTCGATGTGCCGCGGCGCGGTCGGCTCCTGCCCCGCAGACGTTTGACAGACGATTTGGTTCAAAGAACTGAGCACAGAGATACCTGTAAAATGGATCTATAGAGAAAAAAAAAAGTTTGACAACTTTTGACGGTGGATCTCTAGGCTCACGCATCGATGAAGAACGCAGCCAGCTGCGATAAGTAGTGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCAAATGGCGCTCTTGGGTTCTCCCAGGAGCATGTCTGTCTGAGTGTCTATTCAAACACCCATCGTGCGAATCCGCGTGCGCGCGTCCGCCTGGTCCCCGTCTGTAGCGGATGGGGCCGGGACGCGCCCGCCGTGAACGGGTTCGTGCGGCGTTGAGGGTCGCGGTCGAGATGACCGGTCGTGTGCTAGCCTTGGTGCTCGGCCCGACCGGTCGGTTTACCGCGTCCCTTGAAATTCAGGGAGAAAGATCCGCCAAGCGACATCCGGTGTCGCGCGGGGATCATAGAACAAGATACCCTAAACGGCCACGGCTCTATCGCTTTGACGGAACAAATCTTTCTCCATTCGATTCTTTTTTCTGCTGCCGCCGGGTAGCGGTGGCGGCATCTCAAAGGAAAAAAGGAATCGCCTTCCAGACCTCAGATCAGGCAAGGCTACCCGCTGAATTTAAGCATATTAATAAGCGGAGGAAAAGAAACTAACAAGGATTCCCCCAGTAACGGCGAGTGAAGCGGGAAGAGCTCAAATTTGAAATCTCCGATGCTTGCATCGGCGAGTTGTAGTTGCGAGAAGCACTTTCTAGGCGGAGGGGCCCTGCCTAAGTTGCTTGGAACGGCACGTCGCAGAGGGTGACAACCCCGTCTGTGGCAGGGCCGGCCGCCCACGATGTGCTTTCGAAGAGTCGGGTTGTTTGGGAATGCAGCCCAAAATGGGTGGTAAACTCCATCTAAAGCTAAATACCGGCGTGAGACCGATAGCGAACAAGTACCGTGAGGGAAAGATGAAAAGAACTTTGAAAAGAGAGTTAAAAAGTACGTGAAACCGTCAGAAGGGAAACGAATGGACTCAGCAATGCTCCCTTCGAGATTCAGCCGGTCGGCCTGCGCGCGGCTCGGGGTTTGCGGATCCGAACGGACCGCGGTCCCGGGCGCGTGCCTCGCCGCCGTCGCACTTCTCTTGGGCGCGCGTCAGCGTGGGTCGGAACTGGGCGTCGGAGGTGCCGTGGAAGGTAGGTCTGGAGTCTCGGCTCCGGACTGTTACAGCCCGGCTCCCGCGGCTCGGGTCCGACCGAGGACAGTCGCAGCGCGTGCCTCTTGTGGCTAGGGTCCCGTCCTTCCGGCCGGTCGTCGACCGTGGTGGACTGCTTGCAGTGCTCCGCGACTGCTGCCGGTCGTTGGGGCGGTGTTCTCGTACGACGCGCTTGGGACGCTGGCGGTCATATGGGTCCATCCGACCCGTCTTGAAACACGGACCAAGGAGTCTAACATGTGCGCGAGTCTTTGGGTGAGAGAAACCCCGAGGCGCAATGAAAGTGAAGGCGACCTCGTGCCGCCGAGGTGAGATCCGGTCCCCCTTCGGGCGGGGCCGGCGCATCATCGACCGACCTATCCTACCCTTAGGAAGGTTTGAGTGAGAGCGTGTCTGTTGGGACCCGAAAGATGGTGAACTATGCCTGAATAGGGTGAAGCCAGAGGAAACTCTGGTGGAGGCTCGTAGCGATTCTGACGTGCAAATCGATCGTCGAATTTGGGTATAGGGGCGAAAGACTAATCGAACTGTCTAGTAGCTGGTTCCCTCCGAAGTTTCCCTCAGGATAGCTGGCGCTCGGTAAGTGCAGTTTTATCAGGTAAAGCGAATGATTAGAGGCCTTAGGGTCAAAAGGACCTTAACCTATTCTCAAACTTTAAATTGGTAAGATGTCCGACTTGCCCGAATGAAGCCGGACTCGGAATGCGAGTGCCTAGTGGGCCATTTTTGGTAAGCAGAACTGGCGATGCGGGATGAACCGAACGCGGAGTTAAGGTGCCCAAGTCGACGCTCATCAGACCCCACAAAAGGTGTTGGTTGCTATAGACAGCAGGACGGTGGCCATGGAAGTCGGAACCCGCTAAGGAGTGTGTAACAACTCACCTGCCGAAGCAACTAGCCCTGAAAATGGATGGCGCTCAAGCGTCGCACCTATACTCCGCCGTCGGGGCGAGCGGCCTGCCTCGACGAGTAGGAGGGCGCGGGGGTCGTGACGCAGCCTCTGGCGCGAGCCTGGGTGAAACGGCCCCCGGTGCAGATCTTGGTGGTAGTAGCAAATATTCAAATGAGAACTTTGAAGACCGAAGTGGAGAAAGGTTCCATGTGAACAGCAGTTGGACATGGGTTAGTCGATCCTAAGAGATAGGGTAATTCCGTGTCAAAGCGCCCGATGCTCGGGCCGCCTATCGAAAGGGAATCGGGTTAATATTCCCGAACCGGAACGCGGATATTGCCACTCGTTGGCAGGTGCGGCAACGCAACCGAACCCGGAGACGCCGGCGGGGGCCCCGGAAAGAGTTCTCTTTTCTTTTTAACGGACTCTCACCCTGAAATCGGATTGTCCGGAGATAGGGTTCCATGTCCGGTAAAGCACCACACTTCTTGTGGTGTCCGGTGCGCTCTCGACGGCCCTTGAAAATCCGGGGGAGACGATGATTTTCGCGTCCGGTCGTACTCATAACCGCAGCAGGTCTCCAAGGTGAGCAGCCTCTGGTCGATAGAACAATGTAGGTAAGGGAAGTCGGCAAAATGGATCCGTAACTTCGGGAAAAGGATTGGCTCTAAGCGCCGGGTCTCTCGGGCTGAGACTTGAAGCGGTTGGATCCGGCCCGGACTGGCCGTGGTCGAGGGGAGGGGGGGTTCTCTCTCCCTTTCCGAGGTCGCGGTCGGACCGGGAAGGAATCGGCCGTGGATTGGCCCAGCTGTGACCCTCGGGTCAGATCGGGAGGCGACGAACGGCGAACTTAGAACTGGCAGTGACTAGGGGAATCCGACTGTTTAATTAAAACAAAGCATTGCGATGGCCGGAAACGGTGTTGACGCAATGTGATTTCTGCCCAGTGCTCTGAATGTCAAAGTGAAGAAATTCAACCAAGCGCGGGTAAACGGCGGGAGTAACTATGACTCTCTTAAGGTAGCCAAATGCCTCGTCATCTAATTAGTGACGCGCATGAATGGATGAACGAGATTCCCACTGTCCCTATCTACTATCTAGCGAAACCGCAGCAAAGGGAACGGACTTTGCCGAATCAGCGGGGAAAGAAGACCCTGTTGAGCTTGACTCTAGTCTGACCTTGTGAAAAGACATGAGAGGTGTAGAATAAGTGGGAGCAGCTCGCTGCGCCGGTGAAATACCACTACTCTGATCGTTTTTTTACTTATTGGATGGAGCGGAGGCGAACCGCAAGGTTCACTTTCTGGAGTTAAGGCTCCGGCCTCGCGTCGGAGCCGATCCGCGTCCGAGACACCGTCAGGTTGGGAGTTTGGCTGGGGCGGCACATCTGTCAAATGATAACGCAGGTGTCCTAAGGCGAGCTCAGCGAGAACAGAAATCTCGCGTAGAACAAAAGGGTAAAAGCTCGCTTGATTTTGATTTTCAGTAGGAATACAAACCGCGAAAGCGTGGCCTATCGATCCTTTAGTCTTTAGGAGTTTTAAGCTAGAGGTGTCAGAAAAGTTACCACAGGGATAACTGGCTTGTGGCAGCCAAGCGTTCATAGCGACGTTGCTTTTTGATCCTTCGATGTCGGCTCTTCCTATCATTGCGAAGCAGAATTCGCCAAGTGATGGATTGTTCACCCACCAATAGGGAACGTGAGCTGGGTTTAGACCGTCGTGAGACAGGTTAGTTTTACCCTACTGATGAGACGTTGTTGCAATAGTAATTCTGCTCAGTACGAGAGGAACCGCAGATTCAGACCGCTGGCATTTGCACTTGGCTGAAAAGCCAATGGTGCGAAGCTACCATCTGTTGGATTATGACTGAACGCCTCTAAGTCAGAATCCGTGCTAGAAAGCAACGATGATTACCTCCGGTTTACGGTTAGGCGAACAAGAATAGAGGCCCGCTCGTCGGTCCTCGTGCGTCTCCAAGTGCCATGGGGGACAAGAGCCCCCGTTGCACTACCGATAGCCAACTCGAACGTTTCCGGAGATAAATCCTTTGCAGACGACTTAAATAAAGAACGGGGTATTGTAAGTTGTAGAGTAGCCTTGTTGCTACGATCTTCTGAGATTAAGCCTTTGTTCTAAGATTTGTTCCTTTCCAAATCACACATATTTTCATTATAATTGCAAAGTTTTTTTCAACTCCCACGGCCTTTCCCTTTTCCCTTTTCCCTACTCGCCTGCTCGCTTGCTTCCTACCGTCTTACCGTTCCTACCGGGAACGAGCGAGTGCTTGCTGAGGGTTGCCGAGGGAGTTAGTCCGGCAGAGGGCTGCAAAAAAAAAATTCAACTCCCCACAGATGAGACTTTTTTTTTTTTTTGTTAAGTCTCGAGCGTTTTTTTCGATTTTCTCCTTAGAGAGTAGGTGCATCGTACGTAACTTTTCGCGCTGAACACGATGGTGGTATCGCCTCGGGAACTAAAAAAAATTTCCTAACTAAACTTCACCACCATAAACGGCTACAAGGAACTGACCGACGGGCCGCAGACCCATAGATATAGACACAGCCTCTGAAGACCTACTCGCTGGTGCCGACGGTTTTCCATTCCGAGCTGGTTGGAAGGCTCCCCTCCCCAGAGAAAAAGGGGTTTTTGGACCCTGGCCGCGCGGGGATTGGGTCTTAGACGCCGATGTTGCTGCACGTGATGCACAGCGACCAGGCCTAACTCCTGGCGAGCTTAAAGTTCTTGAAAAATCTCGCGAAAGGGCTCTGCTTAGATACAGAAAAACGATTTTTTGGCACTGGCCGCACGGGGATTGGACCCAGGATGCTGATATTCCGGGACGAAGTTGATATCGACCGGTGCTACCTGATGGAGATGTTAAAAAGGTCGAAAAAGGTCGCTAAAGAGCTCTGCTCACCTATCGAAAAGCTGTTTTTAAGCCCTGGCCACGCCGGGATTCACTTGGACTTGGGCTACCTGCTGGTTGTGTTTAAAAGGTCGAAAAC
Step 3: Extracting target amplicon from the region of interest
Now that we know the regions where our A18S-ITS4 amplicon lives in the P. acuta genome assembly and have extracted that region, we can use a tool called, Amplify4 to simulate a PCR run using our region of interest as the template and our A18S and ITS4 primers that we plan to use in situ. The results show the products from the interaction of the primers with the gene. If it is the right gene qPCR should produce a product ~1080 base pairs in length.
PCR product from Amplify4
Our results show that the primers amplified a fragment that is 1098 base pairs in length. This is just about our target size, so it seems that this 1138 base pair region (including the primer annealing area) is our target gene!
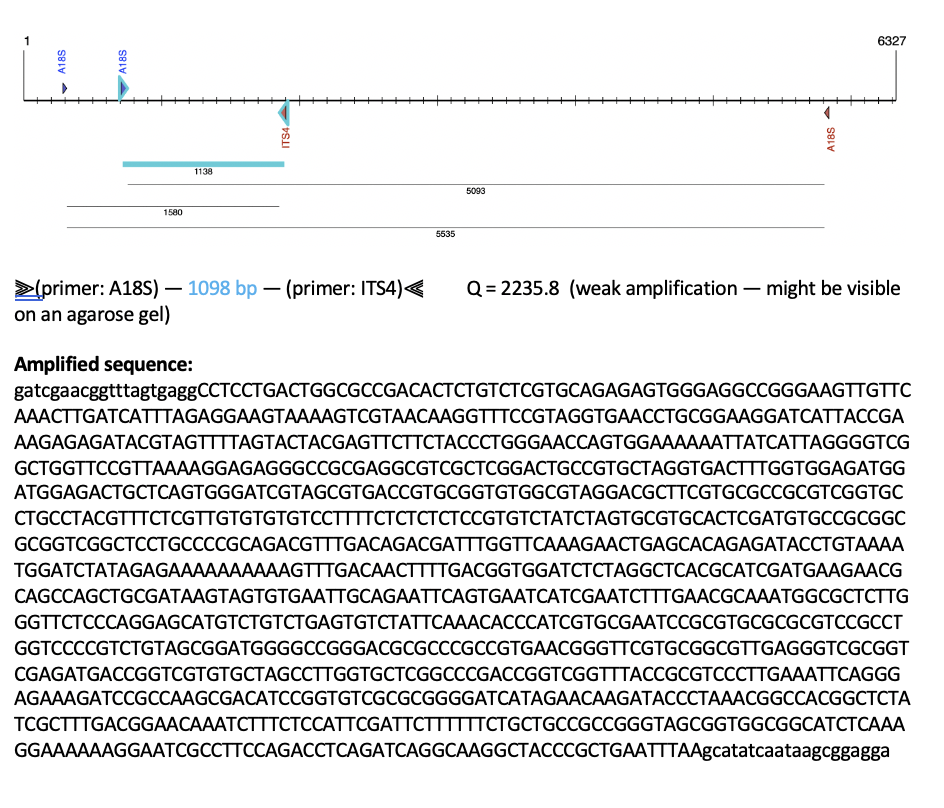
Resulting Amplicon:
>Pocillopora_acuta_HIv2_triploidy_qPCR_standard
GATCGAACGGTTTAGTGAGGCCTCCTGACTGGCGCCGACACTCTGTCTCGTGCAGAGAGTGGGAGGCCGGGAAGTTGTTCAAACTTGATCATTTAGAGGAAGTAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTACCGAAAGAGAGATACGTAGTTTTAGTACTACGAGTTCTTCTACCCTGGGAACCAGTGGAAAAAATTATCATTAGGGGTCGGCTGGTTCCGTTAAAAGGAGAGGGCCGCGAGGCGTCGCTCGGACTGCCGTGCTAGGTGACTTTGGTGGAGATGGATGGAGACTGCTCAGTGGGATCGTAGCGTGACCGTGCGGTGTGGCGTAGGACGCTTCGTGCGCCGCGTCGGTGCCTGCCTACGTTTCTCGTTGTGTGTGTCCTTTTCTCTCTCTCCGTGTCTATCTAGTGCGTGCACTCGATGTGCCGCGGCGCGGTCGGCTCCTGCCCCGCAGACGTTTGACAGACGATTTGGTTCAAAGAACTGAGCACAGAGATACCTGTAAAATGGATCTATAGAGAAAAAAAAAAGTTTGACAACTTTTGACGGTGGATCTCTAGGCTCACGCATCGATGAAGAACGCAGCCAGCTGCGATAAGTAGTGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCAAATGGCGCTCTTGGGTTCTCCCAGGAGCATGTCTGTCTGAGTGTCTATTCAAACACCCATCGTGCGAATCCGCGTGCGCGCGTCCGCCTGGTCCCCGTCTGTAGCGGATGGGGCCGGGACGCGCCCGCCGTGAACGGGTTCGTGCGGCGTTGAGGGTCGCGGTCGAGATGACCGGTCGTGTGCTAGCCTTGGTGCTCGGCCCGACCGGTCGGTTTACCGCGTCCCTTGAAATTCAGGGAGAAAGATCCGCCAAGCGACATCCGGTGTCGCGCGGGGATCATAGAACAAGATACCCTAAACGGCCACGGCTCTATCGCTTTGACGGAACAAATCTTTCTCCATTCGATTCTTTTTTCTGCTGCCGCCGGGTAGCGGTGGCGGCATCTCAAAGGAAAAAAGGAATCGCCTTCCAGACCTCAGATCAGGCAAGGCTACCCGCTGAATTTAAGCATATCAATAAGCGGAGGA
Step 4: Verifying the identity of the amplicon
Next, I verfied that the amplicon corresponds to the ITS region by conducting a quick BLASTN search using the BLAST+ CGI. I did this by copying and pasting the ampified sequence into the query box and typing ‘stony corals (taxid:6125)’ in the optional organism search set parameter.
The BLAST results (link) confirmed that the 104 hits corresponded to the ITS region in Pocillopora spp., and aligned most closely with P. damicornis amplicons. Unfortunately, none of the hits was associated with a publication, so I could not determine if the primers used were A18S and ITS4, like ours. However, all 104 sequences shared >96% identity with our query.
Step 5: Generating a quote
With the identity of our amplicon verified, I navigated to the gBlocks® page IDT website (link) and cliked the “Order Tubes” button. I used “Pocillopora_acuta_HIv2_triploidy_qPCR_standard” as the name and copied our amplicon from Amplify4 into the Sequence box. I then made sure that our sequece passed the complexity test by clicking the “Test Complexity” button. We recieved a score of 5, which was deemed acceptable. Finally, I clicked “Add to Order”, reviewed the Biohazard Disclosure, and sent the quote to purchasing to be ordered!
