Protocol for DNA/RNA Extractions of Montipora Coral Larvae Using Zymo-Duet Extraction Kit
Materials:
- Zymo Duet DNA/RNA Extraction Kit HERE**
- Thermoixer
- Centrifuge capable of 16,000 rcf spin (Eppendorf)**
Make sure ethanol has been added to the wash buffer, and that enzymes have been re-hydrated before starting
Sample Preparation
- Take sample tube with larvae 1 at a time out of the -80 to minimize amount of thawing
- Add 300µl DNA/RNA shield directly to the sample tube
- Record tube number
- Add 30µl of PK digestion buffer to each sample tube
- Add 15µl Proteinase K to each sample tube
- Vortex and spin down sample tubes
- Place in Thermoixer for ~3 hours at 55 degrees C, shaking at 1100 rpm. Check periodically to monitor digestion progress.
- After digestion proceed with DNA and RNA Extraction
DNA Extraction
- Set up yellow DNA spin columns and collection tubes, label appropriately
- Warm elution liquids to 70 degrees C (10mM Tris HCl pH. 8.0 and RNase free water)
- Add equal volume (345µl) DNA/RNA lysis buffer to each sample tube
- Finger flick to mix tubes
- Add 700µl (total volume) of sample gently to the yellow DNA spin column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Important Save the flow through from this step: transfer to a new 1.5mL tube labeled for RNA
- Add 400µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer yellow columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed 10mM Tris HCl to each yellow DNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat steps 18-20 for a final elution volume of 100µl
- Label tubes, store at 4 degrees C if quantifying the same day or the next, if waiting longer store in -20
RNA Extraction
Can do concurrently with DNA Extraction after DNA Extraction Step 7
- Add equal volume (700µl) 100% EtOH to the 1.5mL tubes labeled for RNA containing the original yellow column flow through
- Vortex and spin down to mix
- Add 700µl of that liquid to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Get DNase I from freezer
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Make DNase I treatment master mix:
- 75µl DNA Digestion buffer x # of samples
- 5µl DNase I x # of samples
- Add 80µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubate at room temp for 15 minutes
- Add 400µl DNA/RNA Prep Buffer gently to each column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer green columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed DNase/RNase free water to each green RNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat steps 25-27 for a final elution volume of 100µl
- Label 1.5mL tubes on ice afterwards, and aliquot 5µl into PCR strip tubes to save for Qubit and Tape Station to avoid freeze-thaw of your stock sample
- Store all tubes in the -80
Extraction Content Analysis
These steps analyze the quantity and quality of the DNA/RNA extracted and may be done on a separate day from the extraction
RNA/DNA Quantity
Follow Broad Range dsDNA and RNA Qubit protocol to analyze sample ++quantity++. Read all samples twice.
DNA Quality
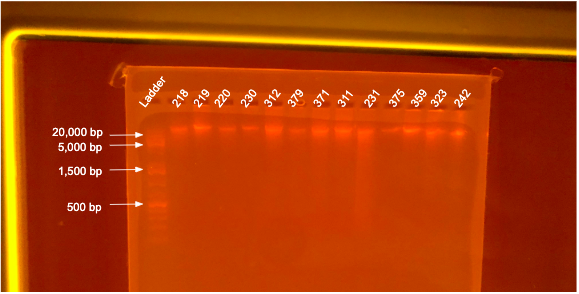
If DNA quantity is sufficient (typically >10 ng/µL) follow the PPP Agarose Gel Protocol to determine DNA quality. “Good” DNA should form a distinct band a the very top of the gel. See example below:
RNA Quality
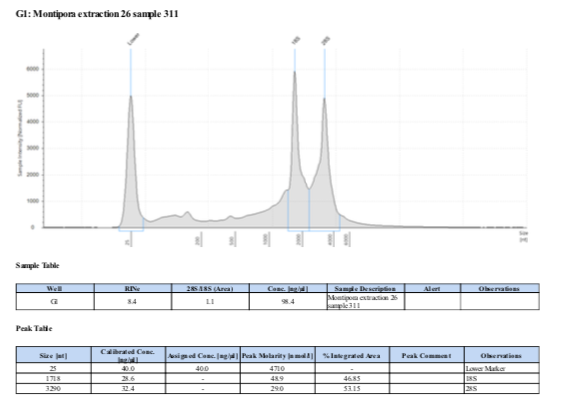
If RNA quantity is sufficient follow the Tape Station Protocol to determine RNA quality and obtain a RNA Integrity Number (RIN). “Good” RNA should have a RIN above 8.0 and form two distinct peaks at the 18S and 28S locations. See example below: